 Over the past decade, teams of engineers, chemists and biologists have analyzed the physical and chemical properties of cicada wings, hoping to unlock the secret to their ability to kill microbes on contact. If this function of nature can be replicated by science, it could lead to products with inherently antibacterial surfaces that are more effective than current chemical treatments.
Over the past decade, teams of engineers, chemists and biologists have analyzed the physical and chemical properties of cicada wings, hoping to unlock the secret to their ability to kill microbes on contact. If this function of nature can be replicated by science, it could lead to products with inherently antibacterial surfaces that are more effective than current chemical treatments.
When researchers in Stony Brook University’s Department of Materials Science and Chemical Engineering developed a simple technique to duplicate the nanostructure of cicada wings, they were still missing a key piece of information. How do the nanopillars on its surface actually kill bacteria? Thankfully, they knew exactly who could help them find the answer: Jan-Michael Carrillo, a researcher in the Center for Nanophase Materials Sciences at the Department of Energy’s Oak Ridge National Laboratory.
For nanoscience researchers seeking computational comparisons and insights for their experiments, Carrillo offers a singular service: large-scale, high-resolution molecular dynamics, or MD, simulations on the Summit supercomputer at the Oak Ridge Leadership Computing Facility at ORNL.
We immediately contacted Jan-Michael and expressed our interest and motivation for the possibility of a simulation. While we know how an MD simulation works, it’s a complicated process and we don’t have much experience doing them, said Maya Endoh, research professor at Stony Brook and co-author of the team’s paper, which was published earlier this year inApplied materials and ACS interfaces.
Getting compute time on Summit isn’t as easy as making a phone call, of course nanoscience researchers must apply to receive such simulation work at the CNMS, and their projects are peer reviewed as part of the application process. But that’s not the only service Carrillo facilitates. In addition to accessing CNMS’s state-of-the-art equipment for nanoscience research, it is also uniquely positioned to help request neutron beam time at ORNL’s Spallation Neutron Source for future experiments.

Our techniques for lipid MD simulations are not unique. What’s unique is that we’ve been able to leverage OLCF resources so we can scan many parameters and build larger systems, Carrillo said. What is also interesting is that ORNLsSNS their techniques match the time scale of MD simulations. So, we plan to compare some of the results from the MD simulations directly with the results in SNS as well as the experiments here in the CNMS.
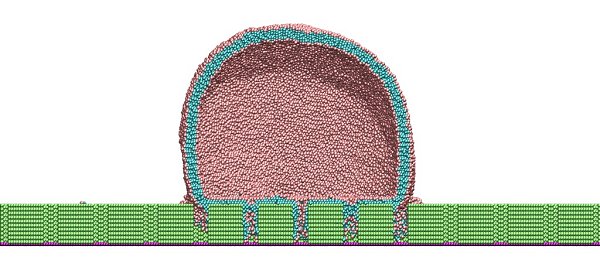
ORNL researchers have simulated the nanostructure of a surface similar to a cicada wing. Side view cross section: Simulated lipid bilayer vesicles interact with nanopillars, showing lipid arrangement and membrane disruption in high curvature regions. Image credit: Jan-Michael Carrillo/ORNL.
Replicate Nature
Stony Brooks Endoh and Tadanori Koga, an associate professor, set out to investigate cicada wings after being inspired by a 2012 research article published in the journal Small that detailed their ability to puncture bacterial cells with lethal results. As researchers in polymeric materials science, Endoh and Koga sought to replicate the wing nanopillars by direct self-assembly.
Self-assembly is a process that uses block copolymers made up of two or more chemically distinct homopolymers that are connected by a covalent bond. The materials offer a simple and effective route to fabricate dense and highly ordered periodic nanostructures with easy control of their geometrical parameters over arbitrarily large areas. For example, the nanopillars on a cicada’s wings are typically 150 nanometers high and apart, but varying those sizes has yielded interesting results.
The cicada wing has a really nice pillar structure, so that’s what we decided to use. But we also wanted to optimize the structure, Koga said. At present we know that the cicada wing can prevent the adhesion of bacteria, but the mechanism is not clear. So, we wanted to control the size and height of the pillar and the distance between the pillars. And then we wanted to see which geometric parameter is crucial for killing bacteria. That’s the whole idea of this project.
Daniel Salatto, a visiting researcher at Brookhaven National Laboratory, was tasked with building the nanosurfaces and conducting experiments on them. To mimic a cicada’s wing, he used a polymer widely used in packaging, specifically a polystyrene-block-poly (methyl methacrylate) block copolymer.
Our original approach to making the bactericidal pillars is very simple: The diblock polymer technically can create the nanostructure on its own as long as we control the environment, Endoh said. We also don’t need a specific type of polymer. That’s why we started with Styrofoam Styrofoam exists everywhere in our daily life. And even if we use a common polymer, we can have the same or similar property which shows the bactericidal property of cicada wing columns.
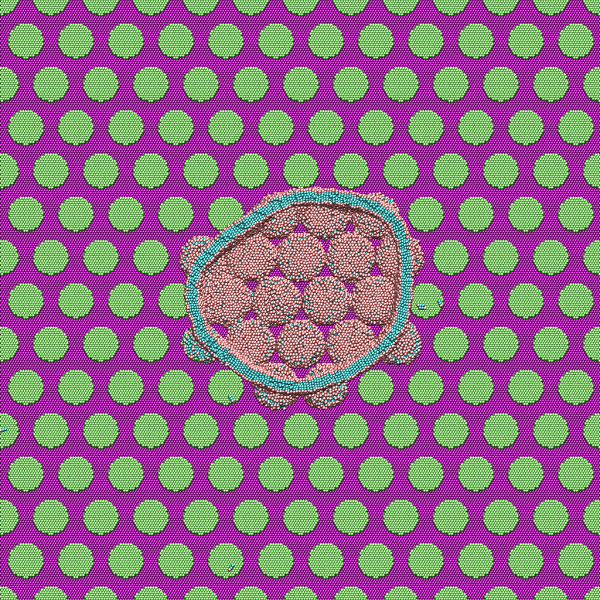
ORNL researchers have simulated the nanostructure of a surface similar to a cicada wing. Top view cross section: Simulated lipid bilayer vesicles interact with nanopillars, showing lipid arrangement and membrane disruption in high curvature regions. Image credit: Jan-Michael Carrillo/ORNL.
Test results experimentally, virtually
Salatto tested the effectiveness of nanosurfaces against bacteria in the laboratory by incubating them in broths of Escherichia coli and Listeria monocytogenes. Once extracted, the samples were examined by fluorescence microscopy and grazing angle X-ray scattering at the Brookhaven Labs National Synchrotron II Light Source to determine what had happened to the bacteria. Not only did the nanosurfaces kill any bacteria that touched them, they also didn’t accumulate any dead bacteria or debris on the surfaces.
It’s known that sometimes when bacterial cells die and adsorb on surfaces, their debris will remain on the surface and thus make a better environment for their siblings to enter and adsorb on top of them, Salatto said. This is where you see a lot of biomedical materials fail, because there’s nothing dealing with debris that works well without using chemicals that more or less could be toxic to the surrounding environments.
But how did the nanosurface pillars achieve this bacterial extermination? This is where Carrillo’s simulations provide some clues to the mystery, showing how and where the bacterium’s cell membrane stretched and collapsed within the pillars’ local structure.
For the Stony Brook project, Carrillo ran an MD simulation consisting of approximately one million particles. The magnitude of the patterns was due to the multiple length scales studied, the size of the lipid molecule and the way it arranges around the nanosurface pillars, the size of the pillars and the length scales of the membrane fluctuations.
The simulation results showed that when there is a strong interaction between the bacterium and the nanosurface substrate, the lipid heads strongly adsorb on the surfaces of the hydrophilic pillars and conform the shape of the membrane to the structure or curvature of the pillars, he said. cart. A stronger attractive interaction further encourages additional adhesion of the membrane to the abutment surfaces. The simulations suggest that membrane rupture occurs when the pillars generate sufficient tension within the lipid bilayer attached to the pillar edges.
This discovery came as a surprise to the Stony Brook team, who expected that closely mimicking nature’s original design would provide the best results. But their best-performing samples didn’t have the same structure or height as the nanopillars of cicada wings.
We thought the height would matter to the nanostructure because we initially expected the height of the pillars to act like a needle to puncture the bacterial membrane. But that’s not how we thought. Even though the height of the nanopillars is short, the bacteria still automatically died, Endoh said. Also, unexpectedly, we didn’t find any absorption on the surface, so it’s self-cleaning. This was thought to be due to the insect flapping its wings to shake off debris. But with our methodology and facilities, we demonstrate that they naturally kill and cleanse themselves.
The team will continue to use simulations to develop a more complete picture of the mechanisms at play, in particular the self-cleaning functionality, before applying the nanosurface to biomedical devices.
As for Carrillo, he will continue his studies of amphiphilic lipid-like bilayer systems, while remaining ready to assist other nanoscience researchers who may need the help of the CNMS, OLCF, or SNS.
Related publication:
D. Salatto et. al., Structure-based design of bactericidal and bacteria-releasing dual nanosurfaces,Applied materials and ACS interfacesJanuary 2023,https://doi.org/10.1021/acsami.2c18121
The CNMS, NSLS-II, OLCF, and SNS are user facilities of the DOE Office of Science. The CNMS integrates nanoscale scientific research with neutron science, synthetic science, and theory/modeling/stimulation. The NSLS-II enables its growing research community to study materials with nanoscale resolution and exquisite sensitivity by providing cutting-edge capabilities. The OLCF provides the computational science community with world-class computing capabilities dedicated to breakthrough science and engineering. The SNS is a new generation spallation neutron source for neutron scattering.
source: Coury Turczyn, science writer, Oak Ridge National Labortory
#OLCF #Stony #Brook #HPC #Simulations #Discover #Cicada #Wings #Kill #Bacteria #Performance #Computing #News #Analysis #insideHPC
Image Source : insidehpc.com
